Lifelike
From Big Data
to Big Picture
Extract concepts easily
Make new connections
Leap beyond traditional research
A graph-powered knowledge mining platform
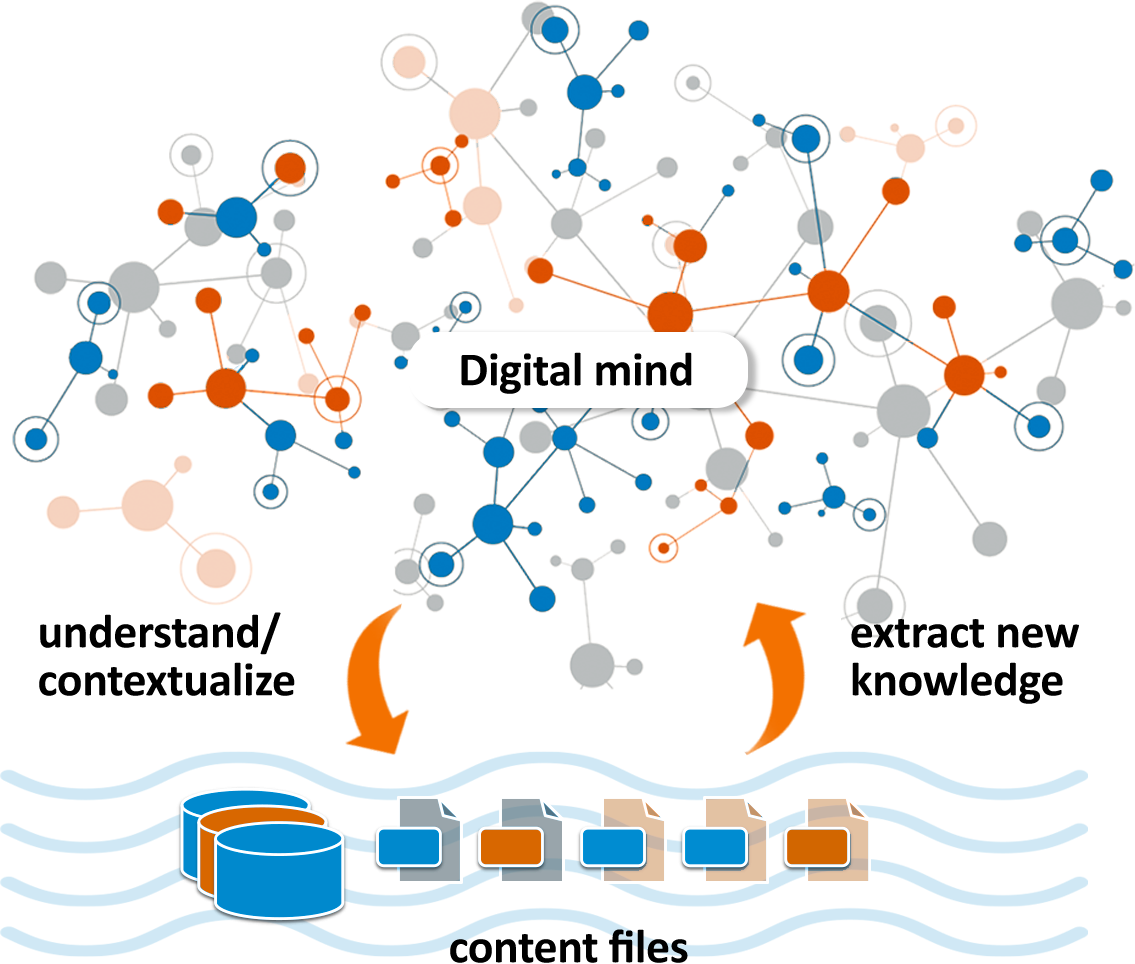
Lifelike has several integrated features that support knowledge mining and collaboration among users in the biosciences field.
Users can upload their own structured and unstructured data. The data is annotated in real-time using NLP and rule-based techniques allowing it to be contextualized and connected to global knowledge. Graph and statistical knowledge mining algorithms can then be run across the combined user and global datasets. In this way users can explore their own data in the broader context of existing knowledge, discovering extended paths comprised of new and previously known relationships.
Lifelike supports communication and collaboration by allowing users to create Knowledge Maps. These are visual graph documents that can be used informally to take "visual notes", or in a more rigorous fashion to create highly curated models that can be analyzed with graph algorithms. Knowledge Maps are a great and easy to use tool to extract knowledge in a visual fashion by simply dragging entities, text snippets, files etc. onto the map. The user can then create new connections between the entities, provide evidence for each connection and add notes and links. The map content is very rich since the entities retain their meta-data and hyperlinks.
Fully integrated with a suite of technologies
Natural Language Processing
Lifelike uses Artificial Intelligence (AI) and rule-based entity recognition to automatically recognize key biological entities such as genes, proteins, diseases, chemicals, phenomena, anatomy, proteins, species and relationships between them. It also enables ‘on the fly’ custom annotations so users can define additional key entities of interest.

Knowledge Reconstruction
Lifelike’s drawing canvas allows users to create connected content and express relationships between biological entities, provide evidence for those relationships including hyperlinks and direct connections to literature and other sources. Drag and drop from annotated papers and enrichment tables allows for fast knowledge reconstruction and visual note taking. Multiple maps can be merged into a comprehensive knowledge graph for graph analysis.
Knowledge Visualization & Enrichment
Lifelike provides an interactive knowledge graph visualizer that clusters relationships by type to simplify complex graphs. Graph nodes can be expanded and detailed supporting data can be viewed. Structured data can be uploaded and enriched against knowledge from many sources. Statistical analysis against ontologies as well as semantic analysis performed on text from public databases enables users to move from big picture view to details.
Search (Full Text and Graph Powered)
Full-text and graph-powered search enables users to find non-obvious connections, and related concepts or find content based on synonyms. This search is seamlessly integrated throughout the application on a button click.